PCA
Download the Rmd notebook for this example
Putting together this page made me realise I still don’t know anything about PCA and factor analysis.
I use the psych package for SPSS-style PCA.
library(tidyverse)
library(psych)
library(viridis)First, I’ll simulate some data with an underlying structure of three factors.
set.seed(444) # for reproducibility; delete when running simulations
a <- rnorm(100, 0, 1)
b <- rnorm(100, 0, 1)
c <- rnorm(100, 0, 1)
df <- data.frame(
id = seq(1,100),
a1 = a + rnorm(100, 0, 1),
a2 = a + rnorm(100, 0, .8),
a3 = a + rnorm(100, 0, .6),
a4 = -a + rnorm(100, 0, .4),
b1 = b + rnorm(100, 0, 1),
b2 = b + rnorm(100, 0, .8),
b3 = b + rnorm(100, 0, .6),
b4 = -b + rnorm(100, 0, .4),
c1 = c + rnorm(100, 0, 1),
c2 = c + rnorm(100, 0, .8),
c3 = c + rnorm(100, 0, .6),
c4 = -c + rnorm(100, 0, .4)
)Select just the columns you want for your PCA. You can visualise their correlations with cor() and ggplot().
traits <- df %>% select(-id)
traits %>%
cor() %>%
as.data.frame() %>%
mutate(var1 = rownames(.)) %>%
gather("var2", "value", a1:c4) %>%
mutate(var1 = factor(var1), var1 = factor(var1, levels = rev(levels(var1)))) %>%
ggplot(aes(var2, var1, fill=value)) +
geom_tile() +
scale_x_discrete(position = "top") +
xlab("") + ylab("") +
scale_fill_viridis(limits=c(-1, 1))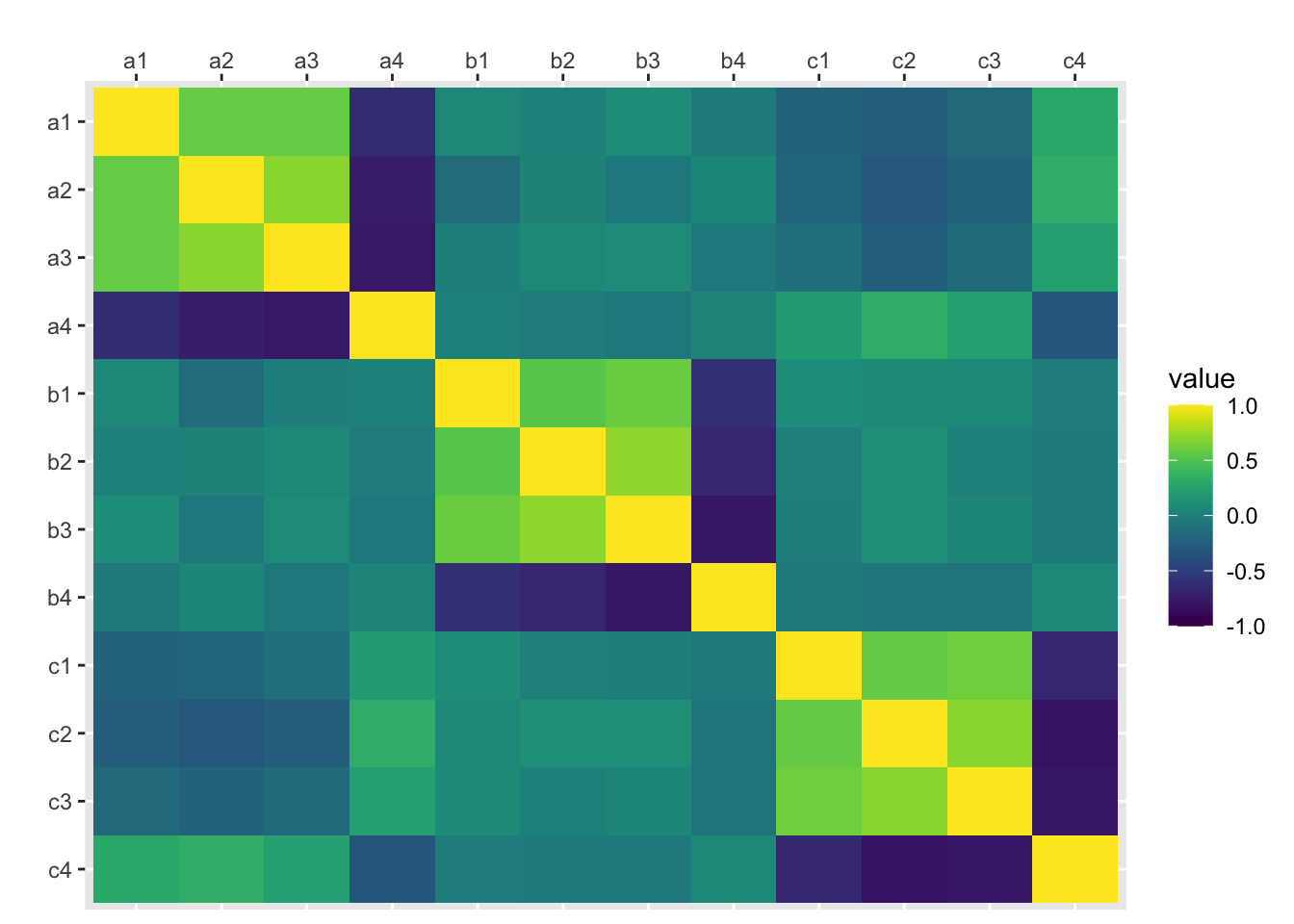
Determine the number of factors to extract. Here I use the SPSS-style default criterion of Eigenvalues > 1
ev <- eigen(cor(traits));
nfactors <- length(ev$values[ev$values > 1]);
nfactors## [1] 3Principal components analysis (SPSS-style)
principal(rotation = “none”)
traits.principal <- principal(traits, nfactors=nfactors, rotate="none", scores=TRUE)
traits.principal## Principal Components Analysis
## Call: principal(r = traits, nfactors = nfactors, rotate = "none", scores = TRUE)
## Standardized loadings (pattern matrix) based upon correlation matrix
## PC1 PC2 PC3 h2 u2 com
## a1 -0.63 0.24 0.42 0.63 0.37 2.1
## a2 -0.72 0.12 0.50 0.77 0.23 1.8
## a3 -0.65 0.26 0.55 0.79 0.21 2.3
## a4 0.73 -0.23 -0.49 0.83 0.17 2.0
## b1 0.14 0.75 -0.19 0.62 0.38 1.2
## b2 0.09 0.83 -0.16 0.71 0.29 1.1
## b3 0.10 0.89 -0.16 0.83 0.17 1.1
## b4 -0.12 -0.88 0.15 0.81 0.19 1.1
## c1 0.64 0.04 0.50 0.66 0.34 1.9
## c2 0.77 0.10 0.42 0.78 0.22 1.6
## c3 0.70 0.08 0.54 0.79 0.21 1.9
## c4 -0.80 -0.04 -0.49 0.88 0.12 1.7
##
## PC1 PC2 PC3
## SS loadings 4.05 3.02 2.03
## Proportion Var 0.34 0.25 0.17
## Cumulative Var 0.34 0.59 0.76
## Proportion Explained 0.45 0.33 0.22
## Cumulative Proportion 0.45 0.78 1.00
##
## Mean item complexity = 1.6
## Test of the hypothesis that 3 components are sufficient.
##
## The root mean square of the residuals (RMSR) is 0.05
## with the empirical chi square 33.59 with prob < 0.44
##
## Fit based upon off diagonal values = 0.98scores.principal <- traits.principal$scorescor(scores.principal, traits) %>%
as.data.frame() %>%
mutate(var1 = rownames(.)) %>%
gather("var2", "value", a1:c4) %>%
mutate(var1 = factor(var1), var1 = factor(var1, levels = rev(levels(var1)))) %>%
ggplot(aes(var2, var1, fill=value)) +
geom_tile() +
scale_x_discrete(position = "top") +
xlab("") + ylab("") +
scale_fill_viridis(limits=c(-1, 1))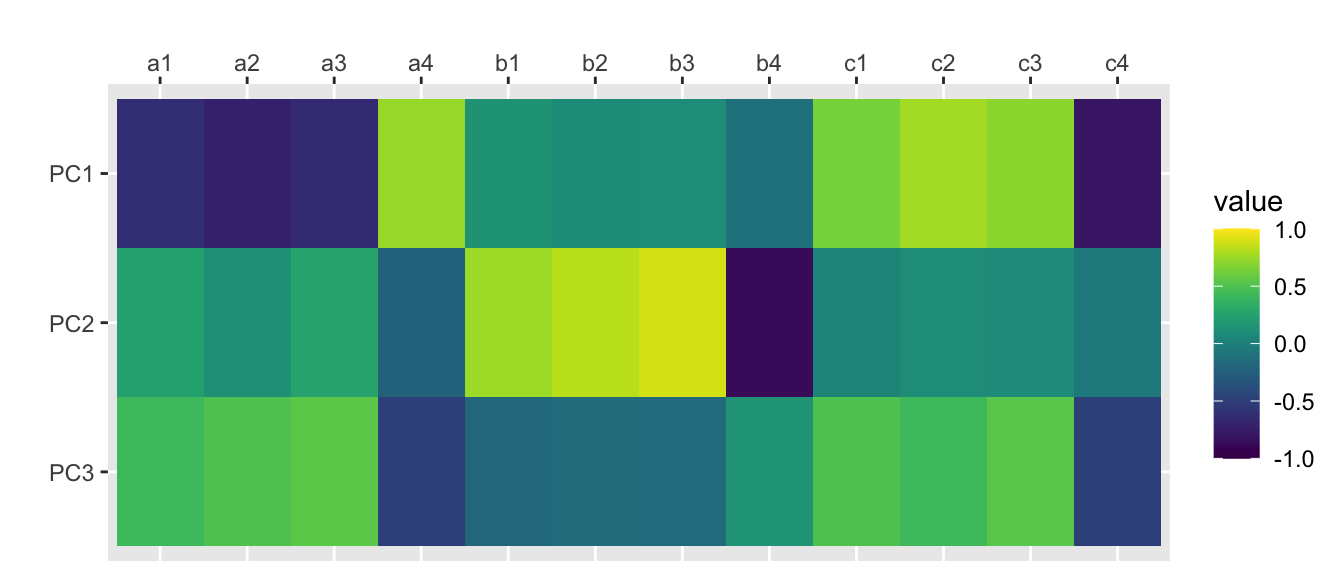
principal(rotation = “varimax”)
traits.varimax <- principal(traits, nfactors=nfactors, rotate="varimax", scores=TRUE)
traits.varimax## Principal Components Analysis
## Call: principal(r = traits, nfactors = nfactors, rotate = "varimax",
## scores = TRUE)
## Standardized loadings (pattern matrix) based upon correlation matrix
## RC1 RC3 RC2 h2 u2 com
## a1 -0.15 0.78 0.06 0.63 0.37 1.1
## a2 -0.16 0.86 -0.09 0.77 0.23 1.1
## a3 -0.07 0.89 0.05 0.79 0.21 1.0
## a4 0.17 -0.90 -0.02 0.83 0.17 1.1
## b1 0.03 -0.05 0.79 0.62 0.38 1.0
## b2 0.02 0.03 0.84 0.71 0.29 1.0
## b3 0.03 0.05 0.91 0.83 0.17 1.0
## b4 -0.05 -0.03 -0.90 0.81 0.19 1.0
## c1 0.81 -0.08 0.00 0.66 0.34 1.0
## c2 0.85 -0.21 0.09 0.78 0.22 1.1
## c3 0.88 -0.09 0.04 0.79 0.21 1.0
## c4 -0.92 0.20 -0.02 0.88 0.12 1.1
##
## RC1 RC3 RC2
## SS loadings 3.09 3.03 2.98
## Proportion Var 0.26 0.25 0.25
## Cumulative Var 0.26 0.51 0.76
## Proportion Explained 0.34 0.33 0.33
## Cumulative Proportion 0.34 0.67 1.00
##
## Mean item complexity = 1
## Test of the hypothesis that 3 components are sufficient.
##
## The root mean square of the residuals (RMSR) is 0.05
## with the empirical chi square 33.59 with prob < 0.44
##
## Fit based upon off diagonal values = 0.98scores.varimax <- traits.varimax$scorescor(scores.varimax, traits) %>%
as.data.frame() %>%
mutate(var1 = rownames(.)) %>%
gather("var2", "value", a1:c4) %>%
mutate(var1 = factor(var1), var1 = factor(var1, levels = rev(levels(var1)))) %>%
ggplot(aes(var2, var1, fill=value)) +
geom_tile() +
scale_x_discrete(position = "top") +
xlab("") + ylab("") +
scale_fill_viridis(limits=c(-1, 1))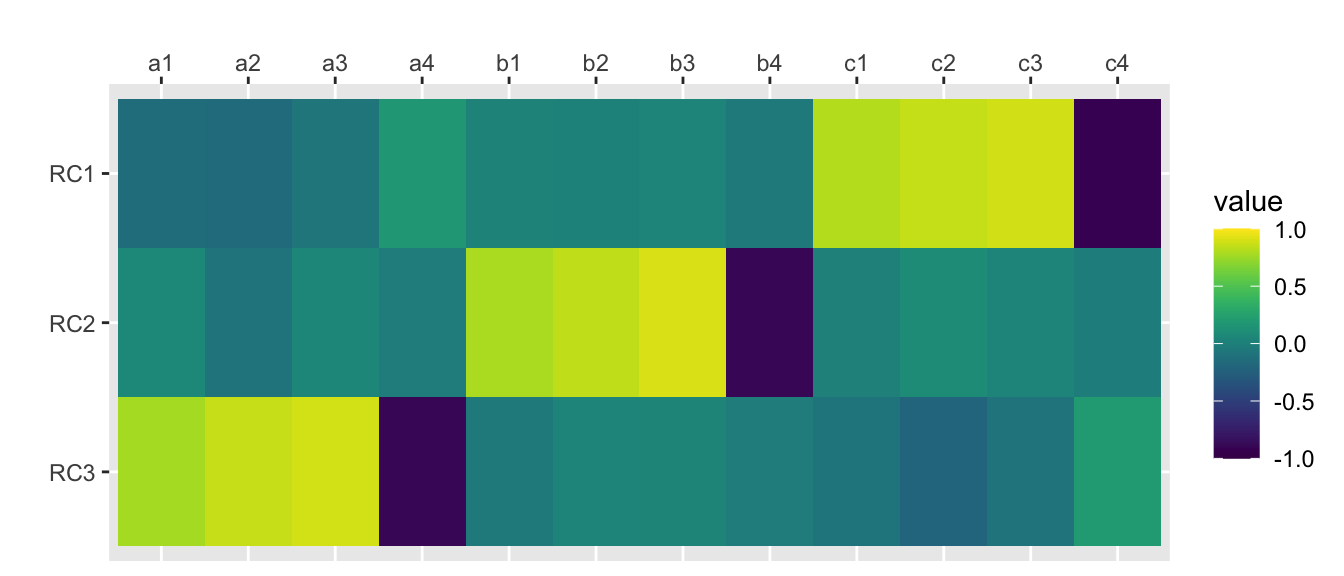
Here are some other functions for PCA/Factor Analysis
princomp()
traits.princomp <- princomp(traits)
traits.princomp$loadings[,1:nfactors]## Comp.1 Comp.2 Comp.3
## a1 0.33006521 0.181709692 0.45906811
## a2 0.29151833 0.067518916 0.38086462
## a3 0.23950762 0.132057875 0.38289101
## a4 -0.25823681 -0.112938603 -0.32926216
## b1 -0.10631062 0.507810878 -0.12639281
## b2 -0.07370167 0.500188081 -0.09771759
## b3 -0.06894907 0.491029731 -0.08032005
## b4 0.07460353 -0.426344256 0.07374535
## c1 -0.44100481 -0.021914387 0.38825383
## c2 -0.42646359 0.010511567 0.23488306
## c3 -0.35757417 -0.001802824 0.29574993
## c4 0.38827130 0.026358392 -0.24163371scores.princomp <- traits.princomp$scores[,1:nfactors]cor(scores.princomp, traits) %>%
as.data.frame() %>%
mutate(var1 = rownames(.)) %>%
gather("var2", "value", a1:c4) %>%
mutate(var1 = factor(var1), var1 = factor(var1, levels = rev(levels(var1)))) %>%
ggplot(aes(var2, var1, fill=value)) +
geom_tile() +
scale_x_discrete(position = "top") +
xlab("") + ylab("") +
scale_fill_viridis(limits=c(-1, 1))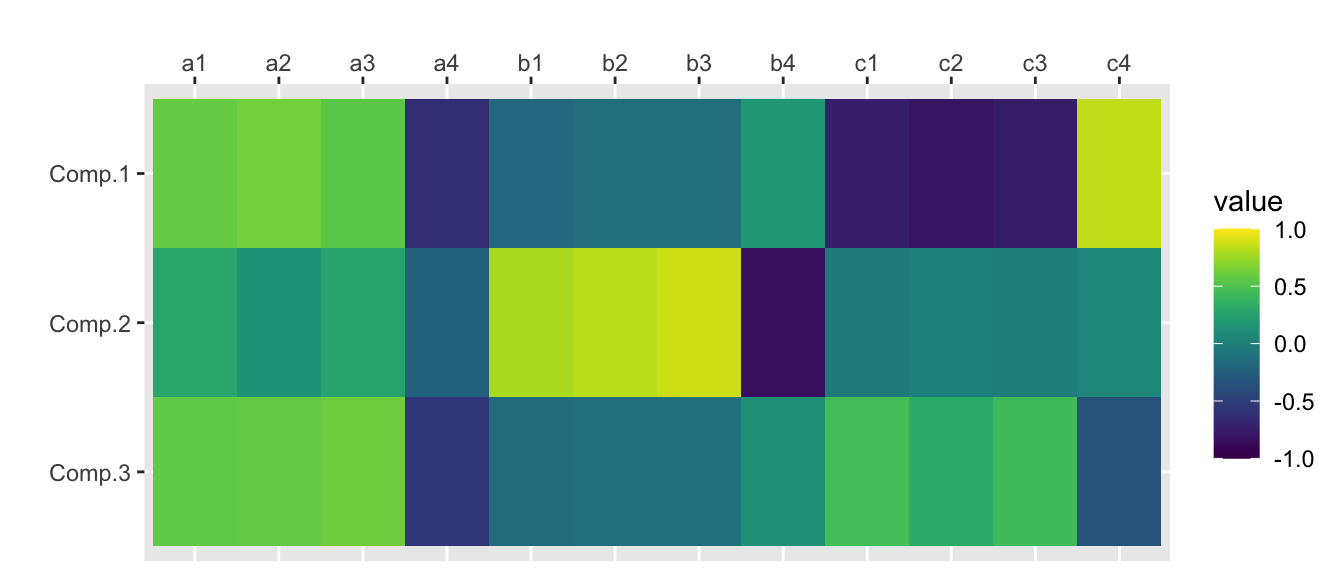
factanal(rotation = “none”)
traits.fa <- factanal(traits, nfactors, rotation="none", scores="regression")
print(traits.fa, digits=2, cutoff=0, sort=FALSE)##
## Call:
## factanal(x = traits, factors = nfactors, scores = "regression", rotation = "none")
##
## Uniquenesses:
## a1 a2 a3 a4 b1 b2 b3 b4 c1 c2 c3 c4
## 0.52 0.32 0.26 0.18 0.53 0.40 0.18 0.23 0.50 0.27 0.32 0.08
##
## Loadings:
## Factor1 Factor2 Factor3
## a1 -0.46 0.26 0.45
## a2 -0.55 0.17 0.60
## a3 -0.47 0.32 0.64
## a4 0.57 -0.30 -0.64
## b1 0.09 0.63 -0.25
## b2 0.07 0.74 -0.21
## b3 0.08 0.87 -0.24
## b4 -0.10 -0.84 0.24
## c1 0.66 0.03 0.25
## c2 0.83 0.09 0.19
## c3 0.77 0.08 0.29
## c4 -0.92 -0.05 -0.28
##
## Factor1 Factor2 Factor3
## SS loadings 3.65 2.71 1.86
## Proportion Var 0.30 0.23 0.16
## Cumulative Var 0.30 0.53 0.68
##
## Test of the hypothesis that 3 factors are sufficient.
## The chi square statistic is 22.21 on 33 degrees of freedom.
## The p-value is 0.923scores.fa <- traits.fa$scorescor(scores.fa, traits) %>%
as.data.frame() %>%
mutate(var1 = rownames(.)) %>%
gather("var2", "value", a1:c4) %>%
mutate(var1 = factor(var1), var1 = factor(var1, levels = rev(levels(var1)))) %>%
ggplot(aes(var2, var1, fill=value)) +
geom_tile() +
scale_x_discrete(position = "top") +
xlab("") + ylab("") +
scale_fill_viridis(limits=c(-1, 1))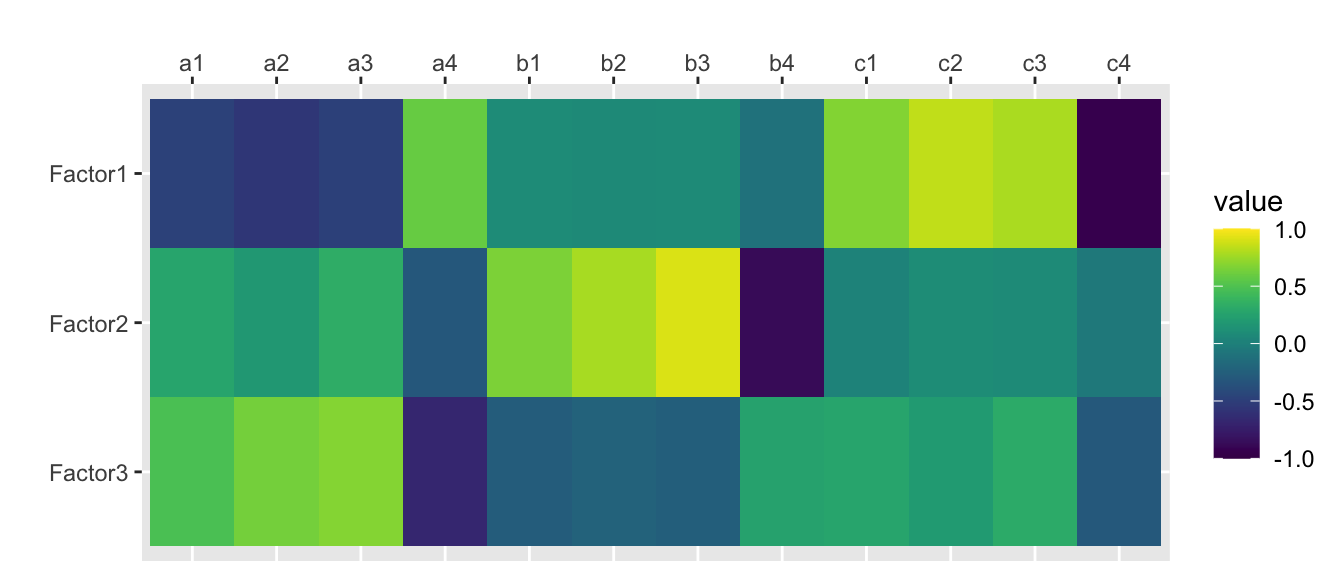
factanal(rotation = “varimax”)
traits.fa.vm <- factanal(traits, nfactors, rotation="varimax", scores="regression")
print(traits.fa.vm, digits=2, cutoff=0, sort=FALSE)##
## Call:
## factanal(x = traits, factors = nfactors, scores = "regression", rotation = "varimax")
##
## Uniquenesses:
## a1 a2 a3 a4 b1 b2 b3 b4 c1 c2 c3 c4
## 0.52 0.32 0.26 0.18 0.53 0.40 0.18 0.23 0.50 0.27 0.32 0.08
##
## Loadings:
## Factor1 Factor2 Factor3
## a1 -0.16 0.67 0.06
## a2 -0.18 0.80 -0.08
## a3 -0.08 0.85 0.05
## a4 0.17 -0.89 -0.03
## b1 0.02 -0.04 0.68
## b2 0.03 0.04 0.77
## b3 0.04 0.05 0.91
## b4 -0.05 -0.03 -0.87
## c1 0.70 -0.10 0.00
## c2 0.82 -0.21 0.09
## c3 0.82 -0.11 0.04
## c4 -0.94 0.19 -0.02
##
## Factor1 Factor2 Factor3
## SS loadings 2.82 2.73 2.67
## Proportion Var 0.23 0.23 0.22
## Cumulative Var 0.23 0.46 0.68
##
## Test of the hypothesis that 3 factors are sufficient.
## The chi square statistic is 22.21 on 33 degrees of freedom.
## The p-value is 0.923scores.fa.vm <- traits.fa.vm$scorescor(scores.fa.vm, traits) %>%
as.data.frame() %>%
mutate(var1 = rownames(.)) %>%
gather("var2", "value", a1:c4) %>%
mutate(var1 = factor(var1), var1 = factor(var1, levels = rev(levels(var1)))) %>%
ggplot(aes(var2, var1, fill=value)) +
geom_tile() +
scale_x_discrete(position = "top") +
xlab("") + ylab("") +
scale_fill_viridis(limits=c(-1, 1))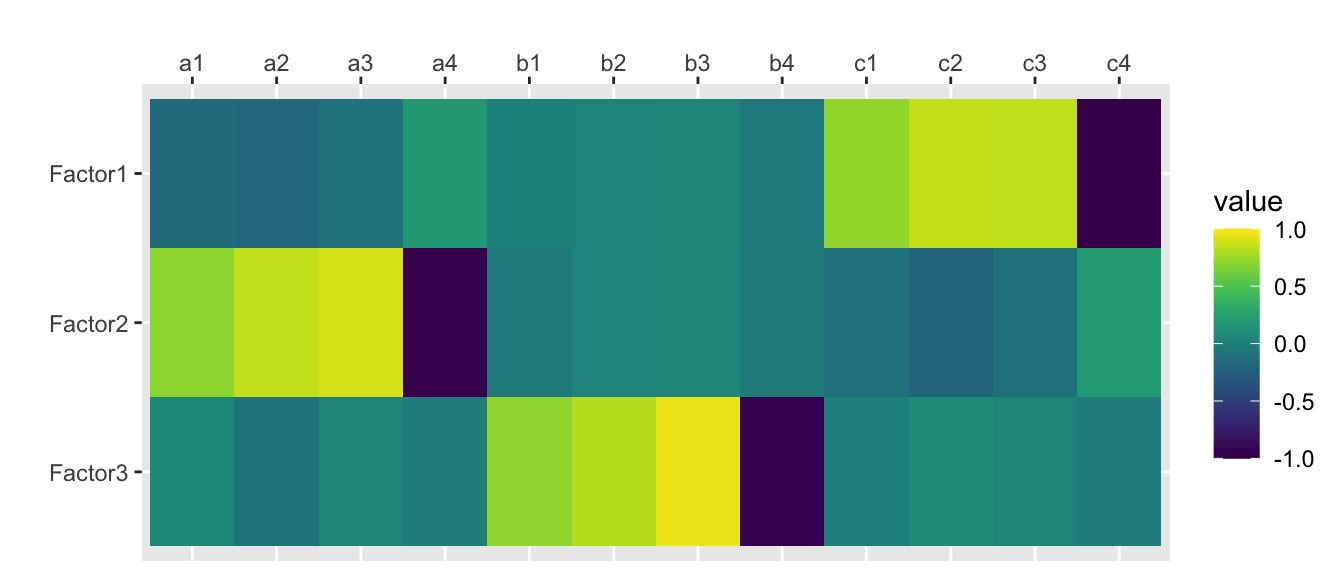
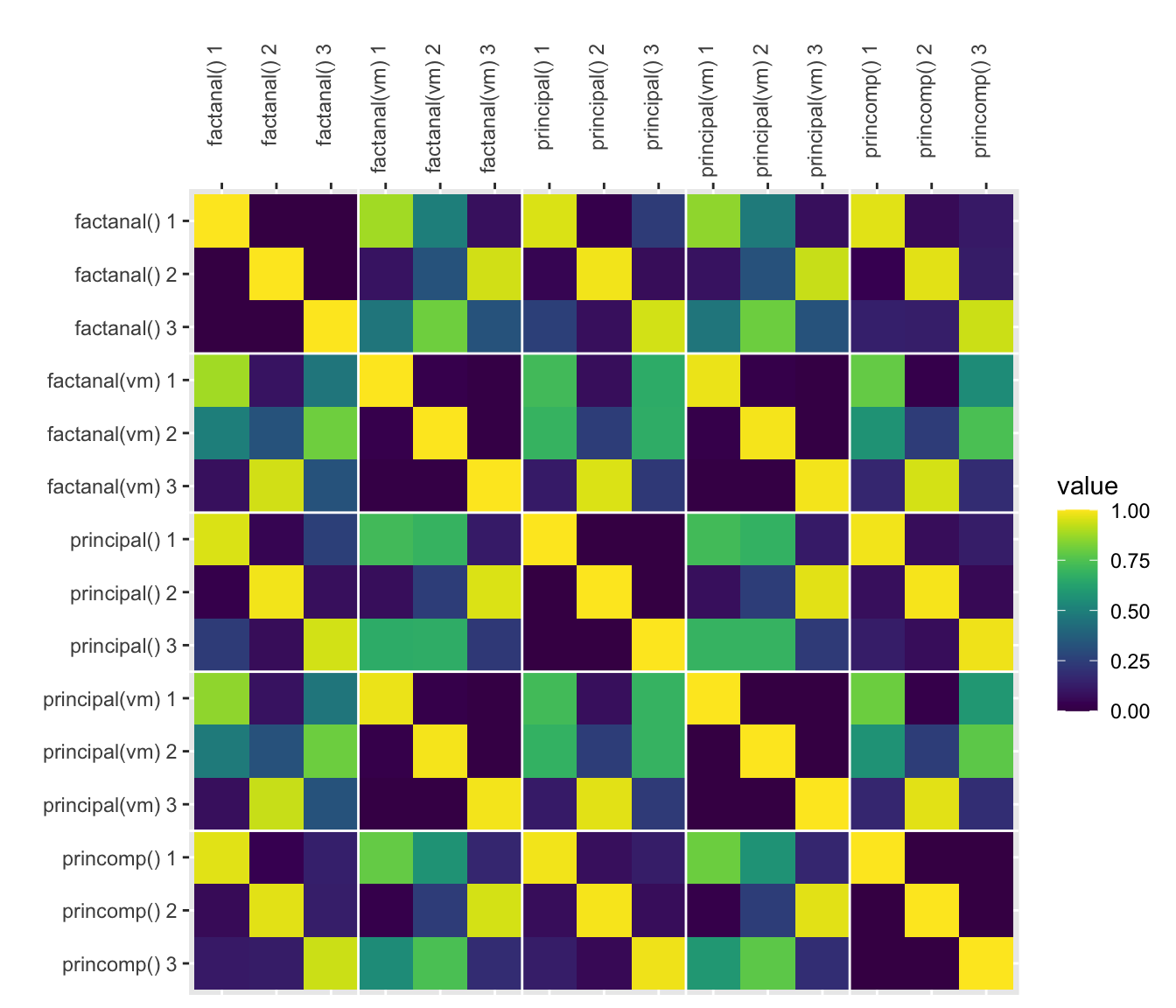
How do they compare?
Here, I’ll plot the absolute value of all the correlations (since the sign on factors/PCs is arbitrary).
The functions principal(rotation = “varimax”) and factanal(rotation = “varimax”) are nearly (but not perfectly) identical.
scores.fa <- traits.fa$scores
colnames(scores.principal) <- c("principal() 1", "principal() 2", "principal() 3")
colnames(scores.varimax) <- c("principal(vm) 1", "principal(vm) 2", "principal(vm) 3")
colnames(scores.princomp) <- c("princomp() 1", "princomp() 2", "princomp() 3")
colnames(scores.fa) <- c("factanal() 1", "factanal() 2", "factanal() 3")
colnames(scores.fa.vm) <- c("factanal(vm) 1", "factanal(vm) 2", "factanal(vm) 3")
cbind(
scores.princomp,
scores.principal,
scores.fa,
scores.varimax,
scores.fa.vm
) %>%
cor() %>%
as.data.frame() %>%
mutate(var1 = rownames(.)) %>%
gather("var2", "value", 1:15) %>%
mutate(var1 = factor(var1), var1 = factor(var1, levels = rev(levels(var1)))) %>%
mutate(value = abs(value)) %>%
ggplot(aes(var2, var1, fill=value)) +
geom_tile() +
scale_x_discrete(position = "top") +
xlab("") + ylab("") +
scale_fill_viridis(limits=c(0, 1)) +
geom_hline(yintercept = 3.5, color="white") +
geom_hline(yintercept = 6.5, color="white") +
geom_hline(yintercept = 9.5, color="white") +
geom_hline(yintercept = 12.5, color="white") +
geom_vline(xintercept = 3.5, color="white") +
geom_vline(xintercept = 6.5, color="white") +
geom_vline(xintercept = 9.5, color="white") +
geom_vline(xintercept = 12.5, color="white") +
theme(axis.text.x=element_text(angle=90,hjust=1))